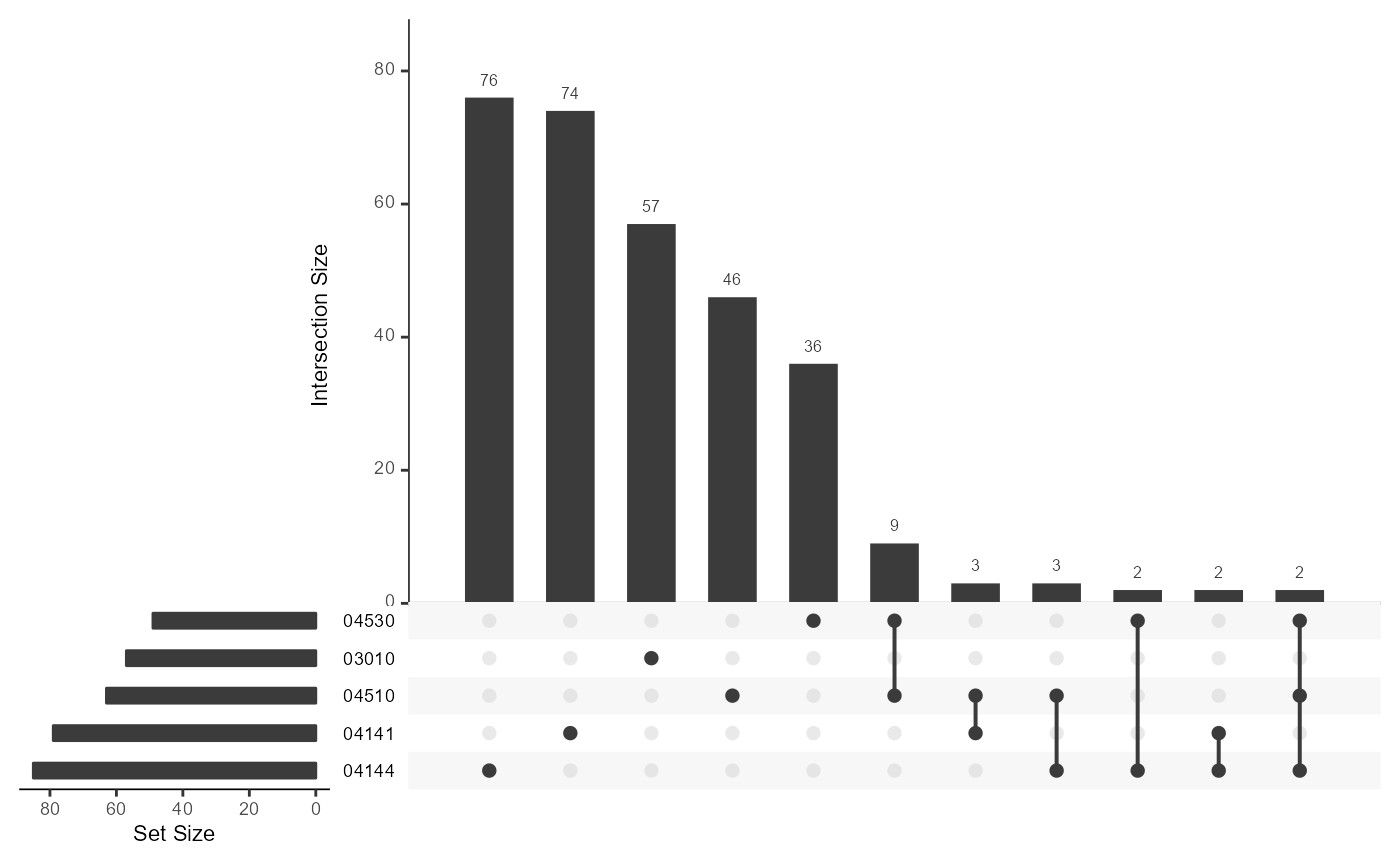
UpSetR wrapper for sigora results
sigora_upsetR.RdUpSetR wrapper for sigora results
sigora_upsetR(sigora_res, GPStable, ...)
Arguments
| sigora_res | Object returned by the |
|---|---|
| GPStable | Object returned by |
| ... | other parameters to |
Examples
sigora_res <- prora::sigora_example GPStable <- prora::GPStab sigora_upsetR(sigora_res, GPStable) #> Joining, by = "pathwayId" #> Joining, by = "gene"df <- get_UniprotID_from_fasta_header(prora::exampleContrastData) myGPSrepo <- makeGPS_wrappR(ids = df$UniprotID) #> 'select()' returned 1:many mapping between keys and columns #> Joining, by = "pathwayID" #> Time difference of 0.674825 secs #> Time difference of 1.163109 secs names(myGPSrepo) #> [1] "gps" "gpsTable" res <- sigoraWrappR(df,score_col = "estimate", GPSrepos = myGPSrepo$gps, threshold = 0.5) #> pathwy.id description pvalues Bonferroni #> 1 03010 Ribosome 1.143e-221 1.532e-219 #> 2 03060 Protein export 1.008e-15 1.351e-13 #> 3 04141 Protein processing in endoplasmic reticulum 4.155e-10 5.568e-08 #> 4 05160 Hepatitis C 2.422e-08 3.245e-06 #> 5 04144 Endocytosis 2.813e-07 3.769e-05 #> 6 04530 Tight junction 2.479e-06 3.322e-04 #> 7 04510 Focal adhesion 3.521e-06 4.718e-04 #> 8 04114 Oocyte meiosis 3.458e-05 4.634e-03 #> 9 04360 Axon guidance 3.606e-05 4.832e-03 #> successes PathwaySize N sample.size #> 1 654.12 1577.52 27971.51 3192.29 #> 2 39.27 83.71 27971.51 3192.29 #> 3 340.68 2176.34 27971.51 3192.29 #> 4 36.99 119.34 27971.51 3192.29 #> 5 343.99 2334.53 27971.51 3192.29 #> 6 82.61 432.19 27971.51 3192.29 #> 7 37.00 149.14 27971.51 3192.29 #> 8 30.88 122.42 27971.51 3192.29 #> 9 27.62 104.61 27971.51 3192.29 sigora_upsetR(res, myGPSrepo$gpsTable) #> Joining, by = "pathwayId" #> Joining, by = "gene"